Home
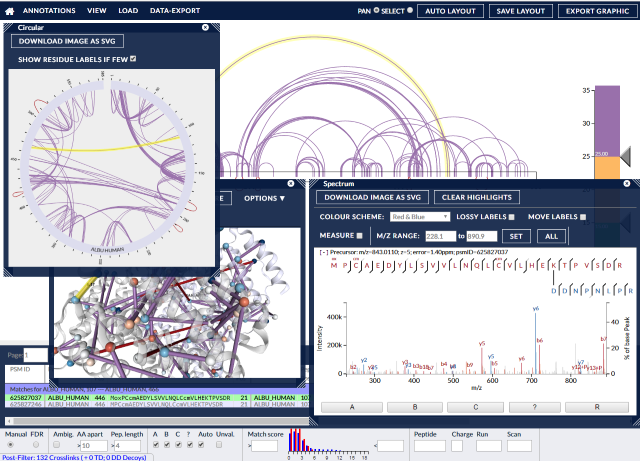
xiVIEW is a web-based visualisation tool for the analysis of cross-linking / mass spectrometry results, it is independent of the search software used. It provides multiple, linked views of the data, including:
- 2D network (xiNET or circular)
- the supporting annotated spectra using xiSPEC.
- 3D structure view using NGL.
The recommended input file format for identifications is version 1.2.0 of mzIdentML. Some CSV formats are also supported.
If your mzIdentML 1.2.0 files work with xiVIEW and spectra are displayed then the files meet the criteria for inclusion in the PRIDE crosslinking archive.
The video tutorials give an overview of xiVIEW's many features.
When using xiVIEW please cite: Graham, M., Combe, C. W., Kolbowski, L. & Rappsilber, J. xiView: A common platform for the downstream analysis of Crosslinking Mass Spectrometry data. doi: 10.1101/561829 .
New User?
Problems? Email administrator

